Barely four months into launching, the core team at Kasturba Hospital’s genome sequencing lab landed their biggest mission. Meet the women playing a critical role in Maharashtra’s fight against COVID’s latest variant
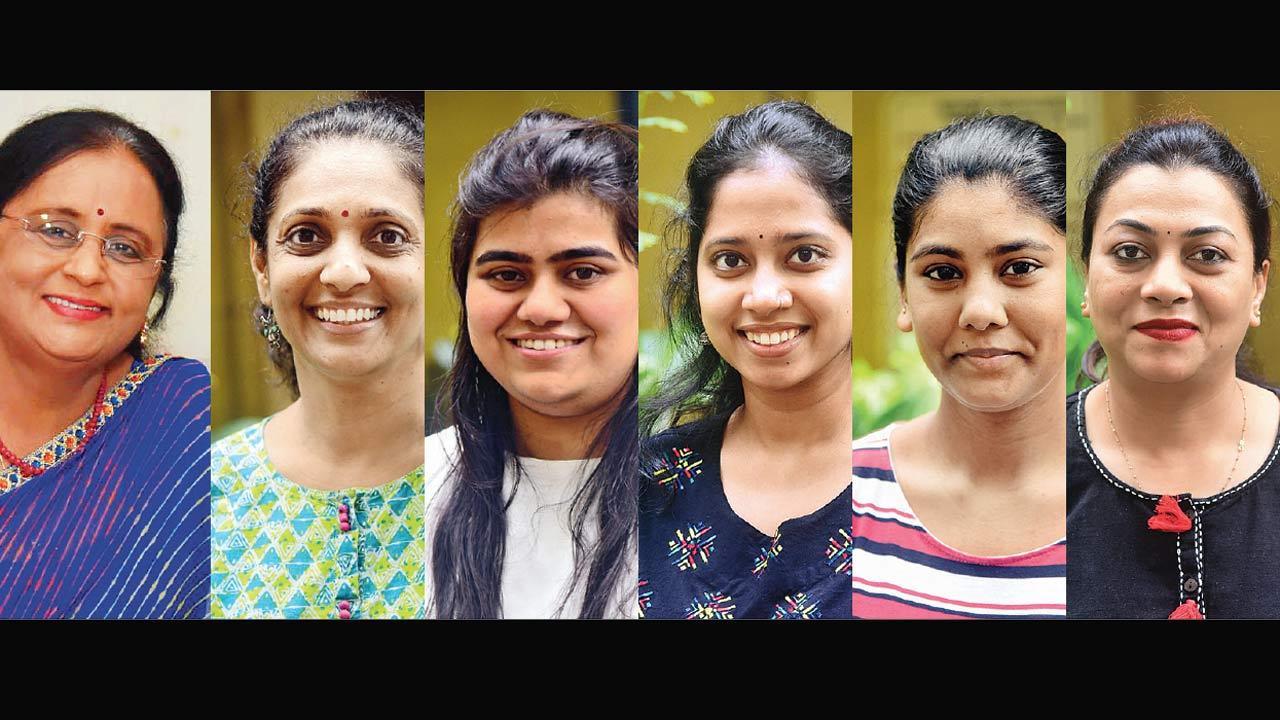
Dr Jayanthi Shastri, laboratory director; Dr Nayana Ingole, deputy lab director; Vidushi Chitalia, research scientist; Sanika Shirsat, research assistant; Reem Nomani, research assistant; Jasmina Savak, research assistant
Dr Jayanthi Shastri, professor and head of the microbiology department at BYL Nair hospital, was in the US last December to visit her children, when a new COVID variant, Alpha (B.1.1.7), was identified in the UK. It was labelled a “variant of concern” by Public Health England. “That’s when it hit me that this was different from the Wuhan strain, and its transmission might lead to enforcement of travel restrictions,” Dr Shastri remembers. She was able to return the next year just in time to battle a double whammy on home turf: community transmission of the Alpha variant in Mumbai followed by the mega outbreak of Delta, which led to the devastating second COVID wave.
Whole Genome Sequencing (WGS) for samples from Maharashtra at the time was being conducted in collaboration with the National Institute of Virology in Pune. “I suggested that we start a WGS lab in Mumbai to the MCGM [Municipal Corporation of Greater Mumbai or Brihanmumbai Municipal Corporation]. We were toying with the idea and trying to put the funds together, when I received an email from Dr Mehul Mehta, chief medical officer at Boston’s Albright Stonebridge Group, and an alumnus of Nair Hospital. He had read about our work at Kasturba Hospital and offered to donate WSG machines from Illumina, a leader in sequencing technology, given that the sequencing technology in Mumbai was abysmal. This was an OMG moment!”
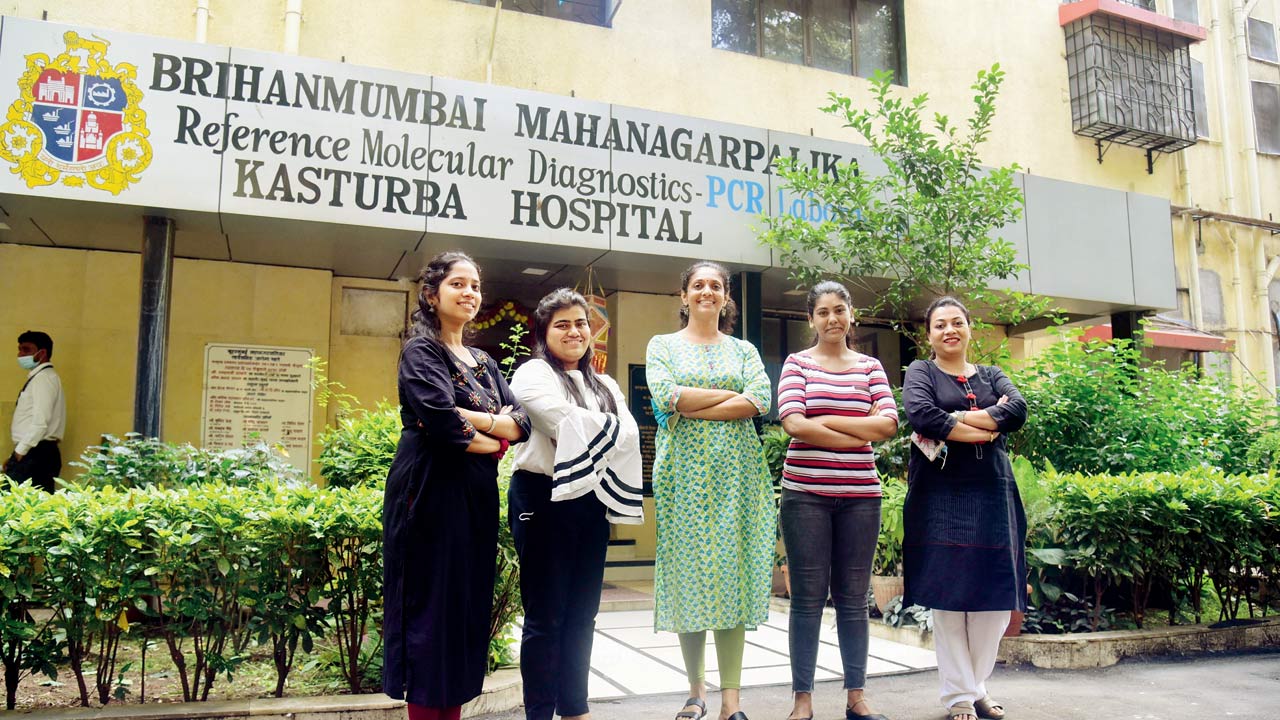 (From left) Sanika Shirsat, Vidushi Chatalia, Dr Nayana Ingole, Reem Nomani and Jasmina Savak at Kasturba Hospital, which houses the sequencing lab. Pics/Shadab Khan
(From left) Sanika Shirsat, Vidushi Chatalia, Dr Nayana Ingole, Reem Nomani and Jasmina Savak at Kasturba Hospital, which houses the sequencing lab. Pics/Shadab Khan
The two high throughput sequencing machines, where multiple viral genomes are analysed from a spectrum of clinical presentations, cost approximately Rs 6.5 crore each.
Mumbai’s first genome sequencing laboratory at Chinchpokli’s Kasturba Hospital for Infectious Diseases is currently processing its sixth batch of positive RT-PCR samples. The machine can process 376 samples at a time. Inaugurated in August this year by Maharashtra Chief Minister Uddhav Thackeray, the infrastructure enables the public health departments to keep an eye out for new variants that could be more infectious, deadlier or in a position to evade vaccines altogether. Until now, Maharashtra had relied on the Indian SARS-CoV-2 Genomic Consortia, or INSACOG, a network of 10 laboratories established by the Union health ministry in December, for genome sequencing. Currently, besides Mumbai, Pune has two genome sequencing labs under the National Institute of Virology and the Council of Scientific and Industrial Research.
 Dr Carlos Ortega of the University of El Salvador Virology Centre works on Coronavirus genome sequencing. More than 1.2 million Coronavirus genome sequences from 172 countries have been shared on GISAID, a global science initiative that provides open-access to genomic data of viruses. Pic/Getty Images
Dr Carlos Ortega of the University of El Salvador Virology Centre works on Coronavirus genome sequencing. More than 1.2 million Coronavirus genome sequences from 172 countries have been shared on GISAID, a global science initiative that provides open-access to genomic data of viruses. Pic/Getty Images
As luck would have it, Dr Shastri is once again stuck in the US after countries imposed travel restrictions following the detection of the new Omicron variant, first identified in South Africa, and now globally seen in more than 20 countries, including India.
But her core team of six is holding fort. The discovery of the new variant of the Coronavirus in Mumbai on December 5—which has a high number of concerning mutations—has kicked off a frenzy of research activity at the lab. “After a sample in the form of the swab is tested positive through the RT-PCR [Reverse transcription-polymerase chain reaction] test, it is sent to us for sequencing. We basically break open the virus’s cells, isolate the genetic material in the form of RNA, from it, convert it into DNA and analyse it using technology,” explains Dr Nayana Ingole, additional professor of microbiology at Topiwala National Medical College (TNMC) and deputy laboratory director of the molecular lab of Kasturba Hospital, who is currently helming the lab. Each round of sequencing takes five days before results can be declared. While access to high throughput DNA sequencing technologies means they can process bulk samples at a time, the drawback is that inputting anything less than the stipulated numbers means wastage and increased costs. The cost of sequencing 384 samples is Rs 10,06,500. As per the government diktat, the turnaround time for sequencing samples of international travellers is 72 hours. “Given that they [international travellers and their contacts] are priority, and we can’t wait for 376 samples to be collected, we send the one or two samples that we receive to NIV Pune. They have the Nanopore sequencing technology, a unique, scalable technology that enables direct, real-time analysis of long DNA or RNA fragments. So they can process the samples faster.” According to experts, the RNA of SARS-CoV-2 contains almost 30,000 molecules with distinct properties.
 Vidushi Chitalia, research scientist
Vidushi Chitalia, research scientist
Dr Ingole says that while the Omicron strain is not reported to cause severe disease, the transmission rate or spread between individuals is relatively high. “Had it not been for genome sequencing, which helped identify the strain, we would have all dismissed it as sardi khaasi. Because it was reported [by South African researchers] and there is wide scale surveillance, we are now aware.” The whole point of WGS, she adds, is timely intervention, and to prevent something from becoming a public health issue. If we would have had this technology earlier, we may have been able to rein in the Delta variant, she thinks.
“When I am undertaking sequencing, I’m not looking to ascertain whether it’s the Alpha, Delta or Omicron variant. It can be any of these variants or even a new one. The objective is to analyse the genetic code of the organism by looking at the molecules. You then compare the final sequence with that of previous strains to check whether it is a new or existing strain,” explains Vidushi Chitalia, research scientist at the Kasturba genome lab. The Ghatkopar resident was involved in research of data devices and has developed RT-PCR and extraction kits, before she was recruited for the job.
 The genome sequencing lab houses two throughput machines, which can process 376 COVID-19 samples at a time
The genome sequencing lab houses two throughput machines, which can process 376 COVID-19 samples at a time
At the lab, the team starts work at 9 am and has an eight-hour shift. While the in-time is fixed, punching out depends on how the day unfolds. The last few weeks have been frenetic, with the team clocking in anywhere between 10 to 14 hours. “Working with a public health department is a different ballgame, and I don’t mean that in a bad way. You need to follow a set of procedures for permissions, but fortunately, the lab is pretty well-coordinated, which makes our job easy.”
For Dr Ingole, who has a teaching job on the side, it’s a tightrope walk. “I can’t say that I won’t go to class because there’s a new Omicron case detected. Everything is a priority.” Research assistants Reema Nomani, Sanika Shirsat and Jasmina Savak were inducted into the team in July this year. They all have a Master of Science degree and molecular testing experience background. “When I was recruited for the job, we had no idea that there would be a new strain and that sequencing would take on such a crucial role in public health.”
 Dr Jayanthi Shastri, professor and head of the microbiology department at BYL Nair hospital and head of the genome sequencing lab at Kasturba Hospital. She says it was with the help of BMC addditional commissioner Suresh Kakani that they were able to iron out the glitches in sourcing the genome sequencing equipment from Singapore
Dr Jayanthi Shastri, professor and head of the microbiology department at BYL Nair hospital and head of the genome sequencing lab at Kasturba Hospital. She says it was with the help of BMC addditional commissioner Suresh Kakani that they were able to iron out the glitches in sourcing the genome sequencing equipment from Singapore
For the team to draw any inferences, they need quality samples. According to Dr Ingole, a good sample is one that has a CT (cycle threshold) value of less than 25. According to the ICMR advisory, the CT value of an RT-PCR reaction refers to the number of cycles after which the virus can be detected. If a higher number of cycles is required, it implies that the virus went undetected when the number of cycles was lower. The lower the CT value, the higher the viral load—because the virus has been spotted after a fewer cycles. “Up to 30 is still acceptable. Ideally, when we sequence it, we should be able to read all the genes present in the organism. If the RNA present in the sample is too little, we get a failure.” The throughput sequencing machine takes 18 hours to process the sample. The next stage of data analysis and interpretation is the most challenging, says Dr Ingole. “I will never label anything Omicron or Delta before confirming it with the NIV. All files are sent to them for confirmation and a quality check before releasing it in the public domain. Let’s say, Omicron has 53 mutations and the sample I have is showing 52. How do I take a call whether it is Omicron? Which is why you need trained bio informationists.”
As per the WHO mandate, data of sequencing done anywhere in the world should be submitted to open-access platforms like GISAID, so that a sequence done in one part of the world can be viewed by someone in another part of the scientific world.
 Research scientist Vidisha Chatalia shows the DRAGEN Server which enables ultra-rapid analysis of data
Research scientist Vidisha Chatalia shows the DRAGEN Server which enables ultra-rapid analysis of data
Dr Shastri, who has been remotely monitoring the working of the lab, says an RT-PCR can only reveal whether the swab sample is positive or negative. To make any more educated guesses about where and how the virus is spreading, you need genome sequencing. “Let’s say there are two outbreaks in Byculla and Mulund. If I have to establish if the two outbreaks are from the same virus strain, I need genome sequencing. Through this, I can find out if it’s the same variant, and even if it’s the same, I can tell what the minor differences between the two [outbreaks] are.”
Her focus now is to bolster efforts with extensive surveillance. Along with the throughput machine from Illumina, she has also received funding to conduct 6,000 genome sequencing tests. “We have exhausted 2,000 until now. Now, I am keen to investigate whether there’s community transmission of Omicron. So the whole of last week, we collected all the positive RT-PCR samples from wards across Mumbai. I’m sequencing all these samples to ascertain whether the new strain is present in any of these samples.” Dr Shastri is now in talks to buy new equipment to ensure the lab doesn’t have to reach out to NIV to sequence smaller samples.
In the meanwhile, it’s business as usual at the lab. Being a small team, they say, has helped in making it a cohesive unit.
Incidentally, Dr Ingole is the oldest in the team. “The girls I work with are as old as my son, which actually makes it more fun.”
13 crore
Cost of two throughput sequencing machines where multiple viral genomes are analysed from a spectrum of clinical presentations
Rs 10 lakh
Approx cost of sequencing 384 COVID positive samples
UK leads the way
The UK has surpassed one million SARS-CoV-2 genome sequences uploaded to the international Global Initiative on Sharing Avian Influenza Data (GISAID) database, meaning the UK represents 24 per cent of all samples uploaded during the pandemic. India has sequenced a meagre 0.2 per cent of its cumulative cases.
 Subscribe today by clicking the link and stay updated with the latest news!" Click here!
Subscribe today by clicking the link and stay updated with the latest news!" Click here!










